DNA methylation

DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
As of 2016, two nucleobases have been found on which natural, enzymatic DNA methylation takes place: adenine and cytosine. The modified bases are N6-methyladenine,[1] 5-methylcytosine[2] and N4-methylcytosine.[3]
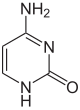
| Unmodified base | 
|
|
||||||
| Adenine, A | Cytosine, C | |||||||
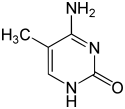
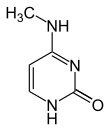
| Modified forms | 
|
|
|
|||||
| N6-Methyladenine, 6mA | 5-Methylcytosine, 5mC | N4-Methylcytosine, 4mC | ||||||
Cytosine methylation is widespread in both eukaryotes and prokaryotes, even though the rate of cytosine DNA methylation can differ greatly between species: 14% of cytosines are methylated in Arabidopsis thaliana, 4% to 8% in Physarum,[4] 7.6% in Mus musculus, 2.3% in Escherichia coli, 0.03% in Drosophila; methylation is essentially undetectable in Dictyostelium;[5][6] and virtually absent (0.0002 to 0.0003%) from Caenorhabditis[7] or fungi such as Saccharomyces cerevisiae and S. pombe (but not N. crassa).[8][9]: 3699 Adenine methylation has been observed in bacterial, plant, and recently in mammalian DNA,[10][11] but has received considerably less attention.
Methylation of cytosine to form 5-methylcytosine occurs at the same 5 position on the pyrimidine ring where the DNA base thymine's methyl group is located; the same position distinguishes thymine from the analogous RNA base uracil, which has no methyl group. Spontaneous deamination of 5-methylcytosine converts it to thymine. This results in a T:G mismatch. Repair mechanisms then correct it back to the original C:G pair; alternatively, they may substitute A for G, turning the original C:G pair into a T:A pair, effectively changing a base and introducing a mutation. This misincorporated base will not be corrected during DNA replication as thymine is a DNA base. If the mismatch is not repaired and the cell enters the cell cycle the strand carrying the T will be complemented by an A in one of the daughter cells, such that the mutation becomes permanent. The near-universal use of thymine exclusively in DNA and uracil exclusively in RNA may have evolved as an error-control mechanism, to facilitate the removal of uracils generated by the spontaneous deamination of cytosine.[12] DNA methylation as well as many of its contemporary DNA methyltransferases have been thought to evolve from early world primitive RNA methylation activity and is supported by several lines of evidence.[13]
In plants and other organisms, DNA methylation is found in three different sequence contexts: CG (or CpG), CHG or CHH (where H correspond to A, T or C). In mammals however, DNA methylation is almost exclusively found in CpG dinucleotides, with the cytosines on both strands being usually methylated. Non-CpG methylation can however be observed in embryonic stem cells,[14][15][16] and has also been indicated in neural development.[17] Furthermore, non-CpG methylation has also been observed in hematopoietic progenitor cells, and it occurred mainly in a CpApC sequence context.[18]
Conserved function of DNA methylation[edit]

The DNA methylation landscape of vertebrates is very particular compared to other organisms. In mammals, around 75% of CpG dinucleotides are methylated in somatic cells,[19] and DNA methylation appears as a default state that has to be specifically excluded from defined locations.[16][20] By contrast, the genome of most plants, invertebrates, fungi, or protists show "mosaic" methylation patterns, where only specific genomic elements are targeted, and they are characterized by the alternation of methylated and unmethylated domains.[21][22]

High CpG methylation in mammalian genomes has an evolutionary cost because it increases the frequency of spontaneous mutations. Loss of amino-groups occurs with a high frequency for cytosines, with different consequences depending on their methylation. Methylated C residues spontaneously deaminate to form T residues over time; hence CpG dinucleotides steadily deaminate to TpG dinucleotides, which is evidenced by the under-representation of CpG dinucleotides in the human genome (they occur at only 21% of the expected frequency).[23] (On the other hand, spontaneous deamination of unmethylated C residues gives rise to U residues, a change that is quickly recognized and repaired by the cell.)
CpG islands[edit]
In mammals, the only exception for this global CpG depletion resides in a specific category of GC- and CpG-rich sequences termed CpG islands that are generally unmethylated and therefore retained the expected CpG content.[24] CpG islands are usually defined as regions with: 1) a length greater than 200bp, 2) a G+C content greater than 50%, 3) a ratio of observed to expected CpG greater than 0.6, although other definitions are sometimes used.[25] Excluding repeated sequences, there are around 25,000 CpG islands in the human genome, 75% of which being less than 850bp long.[23] They are major regulatory units and around 50% of CpG islands are located in gene promoter regions, while another 25% lie in gene bodies, often serving as alternative promoters. Reciprocally, around 60-70% of human genes have a CpG island in their promoter region.[26][27] The majority of CpG islands are constitutively unmethylated and enriched for permissive chromatin modification such as H3K4 methylation. In somatic tissues, only 10% of CpG islands are methylated, the majority of them being located in intergenic and intragenic regions.[citation needed]
Repression of CpG-dense promoters[edit]
DNA methylation was probably present at some extent in very early eukaryote ancestors. In virtually every organism analyzed, methylation in promoter regions correlates negatively with gene expression.[21][28] CpG-dense promoters of actively transcribed genes are never methylated, but, reciprocally, transcriptionally silent genes do not necessarily carry a methylated promoter. In mouse and human, around 60–70% of genes have a CpG island in their promoter region and most of these CpG islands remain unmethylated independently of the transcriptional activity of the gene, in both differentiated and undifferentiated cell types.[29][30] Of note, whereas DNA methylation of CpG islands is unambiguously linked with transcriptional repression, the function of DNA methylation in CG-poor promoters remains unclear; albeit there is little evidence that it could be functionally relevant.[31]
DNA methylation may affect the transcription of genes in two ways. First, the methylation of DNA itself may physically impede the binding of transcriptional proteins to the gene,[32] and second, and likely more important, methylated DNA may be bound by proteins known as methyl-CpG-binding domain proteins (MBDs). MBD proteins then recruit additional proteins to the locus, such as histone deacetylases and other chromatin remodeling proteins that can modify histones, thereby forming compact, inactive chromatin, termed heterochromatin. This link between DNA methylation and chromatin structure is very important. In particular, loss of methyl-CpG-binding protein 2 (MeCP2) has been implicated in Rett syndrome; and methyl-CpG-binding domain protein 2 (MBD2) mediates the transcriptional silencing of hypermethylated genes in "cancer."[citation needed]
Repression of transposable elements[edit]
DNA methylation is a powerful transcriptional repressor, at least in CpG dense contexts. Transcriptional repression of protein-coding genes appears essentially limited to very specific classes of genes that need to be silent permanently and in almost all tissues. While DNA methylation does not have the flexibility required for the fine-tuning of gene regulation, its stability is perfect to ensure the permanent silencing of transposable elements.[33] Transposon control is one of the most ancient functions of DNA methylation that is shared by animals, plants and multiple protists.[34] It is even suggested that DNA methylation evolved precisely for this purpose.[35]
Genome expansion[edit]
DNA methylation of transposable elements has been known to be related to genome expansion. However, the evolutionary driver for genome expansion remains unknown. There is a clear correlation between the size of the genome and CpG, suggesting that the DNA methylation of transposable elements led to a noticeable increase in the mass of DNA.[36]
Methylation of the gene body of highly transcribed genes[edit]
A function that appears even more conserved than transposon silencing is positively correlated with gene expression. In almost all species where DNA methylation is present, DNA methylation is especially enriched in the body of highly transcribed genes.[21][28] The function of gene body methylation is not well understood. A body of evidence suggests that it could regulate splicing[37] and suppress the activity of intragenic transcriptional units (cryptic promoters or transposable elements).[38] Gene-body methylation appears closely tied to H3K36 methylation. In yeast and mammals, H3K36 methylation is highly enriched in the body of highly transcribed genes. In yeast at least, H3K36me3 recruits enzymes such as histone deacetylases to condense chromatin and prevent the activation of cryptic start sites.[39] In mammals, DNMT3a and DNMT3b PWWP domain binds to H3K36me3 and the two enzymes are recruited to the body of actively transcribed genes.[citation needed]
In mammals[edit]

During embryonic development[edit]
DNA methylation patterns are largely erased and then re-established between generations in mammals. Almost all of the methylations from the parents are erased, first during gametogenesis, and again in early embryogenesis, with demethylation and remethylation occurring each time. Demethylation in early embryogenesis occurs in the preimplantation period in two stages – initially in the zygote, then during the first few embryonic replication cycles of morula and blastula. A wave of methylation then takes place during the implantation stage of the embryo, with CpG islands protected from methylation. This results in global repression and allows housekeeping genes to be expressed in all cells. In the post-implantation stage, methylation patterns are stage- and tissue-specific, with changes that would define each individual cell type lasting stably over a long period.[40] Studies on rat limb buds during embryogenesis have further illustrated the dynamic nature of DNA methylation in development. In this context, variations in global DNA methylation were observed across different developmental stages and culture conditions, highlighting the intricate regulation of methylation during organogenesis and its potential implications for regenerative medicine strategies.[41]
Whereas DNA methylation is not necessary per se for transcriptional silencing, it is thought nonetheless to represent a "locked" state that definitely inactivates transcription. In particular, DNA methylation appears critical for the maintenance of mono-allelic silencing in the context of genomic imprinting and X chromosome inactivation.[42][43] In these cases, expressed and silent alleles differ by their methylation status, and loss of DNA methylation results in loss of imprinting and re-expression of Xist in somatic cells. During embryonic development, few genes change their methylation status, at the important exception of many genes specifically expressed in the germline.[44] DNA methylation appears absolutely required in differentiated cells, as knockout of any of the three competent DNA methyltransferase results in embryonic or post-partum lethality. By contrast, DNA methylation is dispensable in undifferentiated cell types, such as the inner cell mass of the blastocyst, primordial germ cells or embryonic stem cells. Since DNA methylation appears to directly regulate only a limited number of genes, how precisely DNA methylation absence causes the death of differentiated cells remain an open question.
Due to the phenomenon of genomic imprinting, maternal and paternal genomes are differentially marked and must be properly reprogrammed every time they pass through the germline. Therefore, during gametogenesis, primordial germ cells must have their original biparental DNA methylation patterns erased and re-established based on the sex of the transmitting parent. After fertilization, the paternal and maternal genomes are once again demethylated and remethylated (except for differentially methylated regions associated with imprinted genes). This reprogramming is likely required for totipotency of the newly formed embryo and erasure of acquired epigenetic changes.[45]
In cancer[edit]
In many disease processes, such as cancer, gene promoter CpG islands acquire abnormal hypermethylation, which results in transcriptional silencing that can be inherited by daughter cells following cell division.[46] Alterations of DNA methylation have been recognized as an important component of cancer development. Hypomethylation, in general, arises earlier and is linked to chromosomal instability and loss of imprinting, whereas hypermethylation is associated with promoters and can arise secondary to gene (oncogene suppressor) silencing, but might be a target for epigenetic therapy.[47] In developmental contexts, dynamic changes in DNA methylation patterns also have significant implications. For instance, in rat limb buds, shifts in methylation status were associated with different stages of chondrogenesis, suggesting a potential link between DNA methylation and the progression of certain developmental processes.[41]
Global hypomethylation has also been implicated in the development and progression of cancer through different mechanisms.[48] Typically, there is hypermethylation of tumor suppressor genes and hypomethylation of oncogenes.[49]
Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation (see DNA methylation in cancer). DNA methylation causing silencing in cancer typically occurs at multiple CpG sites in the CpG islands that are present in the promoters of protein coding genes.[citation needed]
Altered expressions of microRNAs also silence or activate many genes in progression to cancer (see microRNAs in cancer). Altered microRNA expression occurs through hyper/hypo-methylation of CpG sites in CpG islands in promoters controlling transcription of the microRNAs.
Silencing of DNA repair genes through methylation of CpG islands in their promoters appears to be especially important in progression to cancer (see methylation of DNA repair genes in cancer).
In atherosclerosis[edit]
Epigenetic modifications such as DNA methylation have been implicated in cardiovascular disease, including atherosclerosis. In animal models of atherosclerosis, vascular tissue, as well as blood cells such as mononuclear blood cells, exhibit global hypomethylation with gene-specific areas of hypermethylation. DNA methylation polymorphisms may be used as an early biomarker of atherosclerosis since they are present before lesions are observed, which may provide an early tool for detection and risk prevention.[50]
Two of the cell types targeted for DNA methylation polymorphisms are monocytes and lymphocytes, which experience an overall hypomethylation. One proposed mechanism behind this global hypomethylation is elevated homocysteine levels causing hyperhomocysteinemia, a known risk factor for cardiovascular disease. High plasma levels of homocysteine inhibit DNA methyltransferases, which causes hypomethylation. Hypomethylation of DNA affects genes that alter smooth muscle cell proliferation, cause endothelial cell dysfunction, and increase inflammatory mediators, all of which are critical in forming atherosclerotic lesions.[51] High levels of homocysteine also result in hypermethylation of CpG islands in the promoter region of the estrogen receptor alpha (ERα) gene, causing its down regulation.[52] ERα protects against atherosclerosis due to its action as a growth suppressor, causing the smooth muscle cells to remain in a quiescent state.[53] Hypermethylation of the ERα promoter thus allows intimal smooth muscle cells to proliferate excessively and contribute to the development of the atherosclerotic lesion.[54]
Another gene that experiences a change in methylation status in atherosclerosis is the monocarboxylate transporter (MCT3), which produces a protein responsible for the transport of lactate and other ketone bodies out of many cell types, including vascular smooth muscle cells. In atherosclerosis patients, there is an increase in methylation of the CpG islands in exon 2, which decreases MCT3 protein expression. The downregulation of MCT3 impairs lactate transport and significantly increases smooth muscle cell proliferation, which further contributes to the atherosclerotic lesion. An ex vivo experiment using the demethylating agent Decitabine (5-aza-2 -deoxycytidine) was shown to induce MCT3 expression in a dose dependent manner, as all hypermethylated sites in the exon 2 CpG island became demethylated after treatment. This may serve as a novel therapeutic agent to treat atherosclerosis, although no human studies have been conducted thus far.[55]
In heart failure[edit]
In addition to atherosclerosis described above, specific epigenetic changes have been identified in the failing human heart. This may vary by disease etiology. For example, in ischemic heart failure DNA methylation changes have been linked to changes in gene expression that may direct gene expression associated with the changes in heart metabolism known to occur.[56] Additional forms of heart failure (e.g. diabetic cardiomyopathy) and co-morbidities (e.g. obesity) must be explored to see how common these mechanisms are. Most strikingly, in failing human heart these changes in DNA methylation are associated with racial and socioeconomic status which further impact how gene expression is altered,[57] and may influence how the individual's heart failure should be treated.
In aging[edit]
In humans and other mammals, DNA methylation levels can be used to accurately estimate the age of tissues and cell types, forming an accurate epigenetic clock.[58]
A longitudinal study of twin children showed that, between the ages of 5 and 10, there was divergence of methylation patterns due to environmental rather than genetic influences.[59] There is a global loss of DNA methylation during aging.[49]
In a study that analyzed the complete DNA methylomes of CD4+ T cells in a newborn, a 26 years old individual and a 103 years old individual were observed that the loss of methylation is proportional to age.[60] Hypomethylated CpGs observed in the centenarian DNAs compared with the neonates covered all genomic compartments (promoters, intergenic, intronic and exonic regions).[60] However, some genes become hypermethylated with age, including genes for the estrogen receptor, p16, and insulin-like growth factor 2.[49]
In exercise[edit]
High intensity exercise has been shown to result in reduced DNA methylation in skeletal muscle.[61] Promoter methylation of PGC-1α and PDK4 were immediately reduced after high intensity exercise, whereas PPAR-γ methylation was not reduced until three hours after exercise.[61] At the same time, six months of exercise in previously sedentary middle-age men resulted in increased methylation in adipose tissue.[62] One study showed a possible increase in global genomic DNA methylation of white blood cells with more physical activity in non-Hispanics.[63]
In B-cell differentiation[edit]
A study that investigated the methylome of B cells along their differentiation cycle, using whole-genome bisulfite sequencing (WGBS), showed that there is a hypomethylation from the earliest stages to the most differentiated stages. The largest methylation difference is between the stages of germinal center B cells and memory B cells. Furthermore, this study showed that there is a similarity between B cell tumors and long-lived B cells in their DNA methylation signatures.[18]
In the brain[edit]
Two reviews summarize evidence that DNA methylation alterations in brain neurons are important in learning and memory.[64][65] Contextual fear conditioning (a form of associative learning) in animals, such as mice and rats, is rapid and is extremely robust in creating memories.[66] In mice[67] and in rats[68] contextual fear conditioning, within 1–24 hours, it is associated with altered methylations of several thousand DNA cytosines in genes of hippocampus neurons. Twenty four hours after contextual fear conditioning, 9.2% of the genes in rat hippocampus neurons are differentially methylated.[68] In mice,[67] when examined at four weeks after conditioning, the hippocampus methylations and demethylations had been reset to the original naive conditions. The hippocampus is needed to form memories, but memories are not stored there. For such mice, at four weeks after contextual fear conditioning, substantial differential CpG methylations and demethylations occurred in cortical neurons during memory maintenance, and there were 1,223 differentially methylated genes in their anterior cingulate cortex.[67] Mechanisms guiding new DNA methylations and new DNA demethylations in the hippocampus during memory establishment were summarized in 2022.[69] That review also indicated the mechanisms by which the new patterns of methylation gave rise to new patterns of messenger RNA expression. These new messenger RNAs were then transported by messenger RNP particles (neuronal granules) to synapses of the neurons, where they could be translated into proteins.[69] Active changes in neuronal DNA methylation and demethylation appear to act as controllers of synaptic scaling and glutamate receptor trafficking in learning and memory formation.[64]
DNA methyltransferases (in mammals)[edit]

In mammalian cells, DNA methylation occurs mainly at the C5 position of CpG dinucleotides and is carried out by two general classes of enzymatic activities – maintenance methylation and de novo methylation.[70]
Maintenance methylation activity is necessary to preserve DNA methylation after every cellular DNA replication cycle. Without the DNA methyltransferase (DNMT), the replication machinery itself would produce daughter strands that are unmethylated and, over time, would lead to passive demethylation. DNMT1 is the proposed maintenance methyltransferase that is responsible for copying DNA methylation patterns to the daughter strands during DNA replication. Mouse models with both copies of DNMT1 deleted are embryonic lethal at approximately day 9, due to the requirement of DNMT1 activity for development in mammalian cells.[citation needed]
It is thought that DNMT3a and DNMT3b are the de novo methyltransferases that set up DNA methylation patterns early in development. DNMT3L is a protein that is homologous to the other DNMT3s but has no catalytic activity. Instead, DNMT3L assists the de novo methyltransferases by increasing their ability to bind to DNA and stimulating their activity. Mice and rats have a third functional de novo methyltransferase enzyme named DNMT3C, which evolved as a paralog of Dnmt3b by tandem duplication in the common ancestral of Muroidea rodents. DNMT3C catalyzes the methylation of promoters of transposable elements during early spermatogenesis, an activity shown to be essential for their epigenetic repression and male fertility.[71][72] It is yet unclear if in other mammals that do not have DNMT3C (like humans) rely on DNMT3B or DNMT3A for de novo methylation of transposable elements in the germline. Finally, DNMT2 (TRDMT1) has been identified as a DNA methyltransferase homolog, containing all 10 sequence motifs common to all DNA methyltransferases; however, DNMT2 (TRDMT1) does not methylate DNA but instead methylates cytosine-38 in the anticodon loop of aspartic acid transfer RNA.[73]
Since many tumor suppressor genes are silenced by DNA methylation during carcinogenesis, there have been attempts to re-express these genes by inhibiting the DNMTs. 5-Aza-2'-deoxycytidine (decitabine) is a nucleoside analog that inhibits DNMTs by trapping them in a covalent complex on DNA by preventing the β-elimination step of catalysis, thus resulting in the enzymes' degradation. However, for decitabine to be active, it must be incorporated into the genome of the cell, which can cause mutations in the daughter cells if the cell does not die. In addition, decitabine is toxic to the bone marrow, which limits the size of its therapeutic window. These pitfalls have led to the development of antisense RNA therapies that target the DNMTs by degrading their mRNAs and preventing their translation. However, it is currently unclear whether targeting DNMT1 alone is sufficient to reactivate tumor suppressor genes silenced by DNA methylation.[citation needed]
In plants[edit]
Significant progress has been made in understanding DNA methylation in the model plant Arabidopsis thaliana. DNA methylation in plants differs from that of mammals: while DNA methylation in mammals mainly occurs on the cytosine nucleotide in a CpG site, in plants the cytosine can be methylated at CpG, CpHpG, and CpHpH sites, where H represents any nucleotide but not guanine.[74] Overall, Arabidopsis DNA is highly methylated, mass spectrometry analysis estimated 14% of cytosines to be modified.[9]: abstract Later, bisulfite sequencing data estimated that around 25% of Arabidopsis CG sites are methylated, but these levels vary based on the geographic location of Arabidopsis accessions (plants in the north are more highly methylated than southern accessions).[75]
The principal Arabidopsis DNA methyltransferase enzymes, which transfer and covalently attach methyl groups onto DNA, are DRM2, MET1, and CMT3. Both the DRM2 and MET1 proteins share significant homology to the mammalian methyltransferases DNMT3 and DNMT1, respectively, whereas the CMT3 protein is unique to the plant kingdom. There are currently two classes of DNA methyltransferases: 1) the de novo class or enzymes that create new methylation marks on the DNA; 2) a maintenance class that recognizes the methylation marks on the parental strand of DNA and transfers new methylation to the daughter strands after DNA replication. DRM2 is the only enzyme that has been implicated as a de novo DNA methyltransferase. DRM2 has also been shown, along with MET1 and CMT3 to be involved in maintaining methylation marks through DNA replication.[76] Other DNA methyltransferases are expressed in plants but have no known function (see the Chromatin Database).
Genome-wide levels of DNA methylation vary widely between plant species, and Arabidopsis cytosines tend to be less densely methylated than those in other plants. For example, ~92.5% of CpG cytosines are methylated in Beta vulgaris.[77] The patterns of methylation also differ between cytosine sequence contexts; universally, CpG methylation is higher than CHG and CHH methylation, and CpG methylation can be found in both active genes and transposable elements, while CHG and CHH are usually relegated to silenced transposable elements.[78][74]
It is not clear how the cell determines the locations of de novo DNA methylation, but evidence suggests that for many (though not all) locations, RNA-directed DNA methylation (RdDM) is involved. In RdDM, specific RNA transcripts are produced from a genomic DNA template, and this RNA forms secondary structures called double-stranded RNA molecules.[79] The double-stranded RNAs, through either the small interfering RNA (siRNA) or microRNA (miRNA) pathways direct de-novo DNA methylation of the original genomic location that produced the RNA.[79] This sort of mechanism is thought to be important in cellular defense against RNA viruses and/or transposons, both of which often form a double-stranded RNA that can be mutagenic to the host genome. By methylating their genomic locations, through an as yet poorly understood mechanism, they are shut off and are no longer active in the cell, protecting the genome from their mutagenic effect. Recently, it was described that methylation of the DNA is the main determinant of embryogenic cultures formation from explants in woody plants and is regarded the main mechanism that explains the poor response of mature explants to somatic embryogenesis in the plants (Isah 2016).[citation needed]
In insects[edit]
Diverse orders of insects show varied patterns of DNA methylation, from almost undetectable levels in flies to low levels in butterflies and higher in true bugs and some cockroaches (up to 14% of all CG sites in Blattella asahinai).[80]
Functional DNA methylation has been discovered in Honey Bees.[81][82] DNA methylation marks are mainly on the gene body, and current opinions on the function of DNA methylation is gene regulation via alternative splicing[83]
DNA methylation levels in Drosophila melanogaster are nearly undetectable.[84] Sensitive methods applied to Drosophila DNA Suggest levels in the range of 0.1–0.3% of total cytosine.[85] This low level of methylation[86] appears to reside in genomic sequence patterns that are very different from patterns seen in humans, or in other animal or plant species to date. Genomic methylation in D. melanogaster was found at specific short motifs (concentrated in specific 5-base sequence motifs that are CA- and CT-rich but depleted of guanine) and is independent of DNMT2 activity. Further, highly sensitive mass spectrometry approaches,[87] have now demonstrated the presence of low (0.07%) but significant levels of adenine methylation during the earliest stages of Drosophila embryogenesis.
In fungi[edit]
Many fungi have low levels (0.1 to 0.5%) of cytosine methylation, whereas other fungi have as much as 5% of the genome methylated.[88] This value seems to vary both among species and among isolates of the same species.[89] There is also evidence that DNA methylation may be involved in state-specific control of gene expression in fungi.[citation needed] However, at a detection limit of 250 attomoles by using ultra-high sensitive mass spectrometry DNA methylation was not confirmed in single cellular yeast species such as Saccharomyces cerevisiae or Schizosaccharomyces pombe, indicating that yeasts do not possess this DNA modification.[9]: abstract
Although brewers' yeast (Saccharomyces), fission yeast (Schizosaccharomyces), and Aspergillus flavus[90] have no detectable DNA methylation, the model filamentous fungus Neurospora crassa has a well-characterized methylation system.[91] Several genes control methylation in Neurospora and mutation of the DNA methyl transferase, dim-2, eliminates all DNA methylation but does not affect growth or sexual reproduction. While the Neurospora genome has very little repeated DNA, half of the methylation occurs in repeated DNA including transposon relics and centromeric DNA. The ability to evaluate other important phenomena in a DNA methylase-deficient genetic background makes Neurospora an important system in which to study DNA methylation.[citation needed]
In other eukaryotes[edit]
DNA methylation is largely absent from Dictyostelium discoidium[92] where it appears to occur at about 0.006% of cytosines.[6] In contrast, DNA methylation is widely distributed in Physarum polycephalum[93] where 5-methylcytosine makes up as much as 8% of total cytosine[4]
In bacteria[edit]

Adenine or cytosine methylation are mediated by restriction modification systems of many bacteria, in which specific DNA sequences are methylated periodically throughout the genome.[95] A methylase is the enzyme that recognizes a specific sequence and methylates one of the bases in or near that sequence. Foreign DNAs (which are not methylated in this manner) that are introduced into the cell are degraded by sequence-specific restriction enzymes and cleaved. Bacterial genomic DNA is not recognized by these restriction enzymes. The methylation of native DNA acts as a sort of primitive immune system, allowing the bacteria to protect themselves from infection by bacteriophage.[citation needed]
E. coli DNA adenine methyltransferase (Dam) is an enzyme of ~32 kDa that does not belong to a restriction/modification system. The target recognition sequence for E. coli Dam is GATC, as the methylation occurs at the N6 position of the adenine in this sequence (G meATC). The three base pairs flanking each side of this site also influence DNA–Dam binding. Dam plays several key roles in bacterial processes, including mismatch repair, the timing of DNA replication, and gene expression. As a result of DNA replication, the status of GATC sites in the E. coli genome changes from fully methylated to hemimethylated. This is because adenine introduced into the new DNA strand is unmethylated. Re-methylation occurs within two to four seconds, during which time replication errors in the new strand are repaired. Methylation, or its absence, is the marker that allows the repair apparatus of the cell to differentiate between the template and nascent strands. It has been shown that altering Dam activity in bacteria results in an increased spontaneous mutation rate. Bacterial viability is compromised in dam mutants that also lack certain other DNA repair enzymes, providing further evidence for the role of Dam in DNA repair.
One region of the DNA that keeps its hemimethylated status for longer is the origin of replication, which has an abundance of GATC sites. This is central to the bacterial mechanism for timing DNA replication. SeqA binds to the origin of replication, sequestering it and thus preventing methylation. Because hemimethylated origins of replication are inactive, this mechanism limits DNA replication to once per cell cycle.
Expression of certain genes, for example, those coding for pilus expression in E. coli, is regulated by the methylation of GATC sites in the promoter region of the gene operon. The cells' environmental conditions just after DNA replication determine whether Dam is blocked from methylating a region proximal to or distal from the promoter region. Once the pattern of methylation has been created, the pilus gene transcription is locked in the on or off position until the DNA is again replicated. In E. coli, these pili operons have important roles in virulence in urinary tract infections. It has been proposed[by whom?] that inhibitors of Dam may function as antibiotics.
On the other hand, DNA cytosine methylase targets CCAGG and CCTGG sites to methylate cytosine at the C5 position (C meC(A/T) GG). The other methylase enzyme, EcoKI, causes methylation of adenines in the sequences AAC(N6)GTGC and GCAC(N6)GTT.
In Clostridioides difficile, DNA methylation at the target motif CAAAAA was shown to impact sporulation, a key step in disease transmission, as well as cell length, biofilm formation and host colonization.[96]
Molecular cloning[edit]
Most strains used by molecular biologists are derivatives of E. coli K-12, and possess both Dam and Dcm, but there are commercially available strains that are dam-/dcm- (lack of activity of either methylase). In fact, it is possible to unmethylate the DNA extracted from dam+/dcm+ strains by transforming it into dam-/dcm- strains. This would help digest sequences that are not being recognized by methylation-sensitive restriction enzymes.[97][98]
The restriction enzyme DpnI can recognize 5'-GmeATC-3' sites and digest the methylated DNA. Being such a short motif, it occurs frequently in sequences by chance, and as such its primary use for researchers is to degrade template DNA following PCRs (PCR products lack methylation, as no methylases are present in the reaction). Similarly, some commercially available restriction enzymes are sensitive to methylation at their cognate restriction sites and must as mentioned previously be used on DNA passed through a dam-/dcm- strain to allow cutting.[citation needed]
Detection[edit]
DNA methylation can be detected by the following assays currently used in scientific research:[99]
- Mass spectrometry is a very sensitive and reliable analytical method to detect DNA methylation. MS, in general, is however not informative about the sequence context of the methylation, thus limited in studying the function of this DNA modification.
- Methylation-Specific PCR (MSP), which is based on a chemical reaction of sodium bisulfite with DNA that converts unmethylated cytosines of CpG dinucleotides to uracil or UpG, followed by traditional PCR.[100] However, methylated cytosines will not be converted in this process, and primers are designed to overlap the CpG site of interest, which allows one to determine methylation status as methylated or unmethylated.
- Whole genome bisulfite sequencing, also known as BS-Seq, which is a high-throughput genome-wide analysis of DNA methylation. It is based on the aforementioned sodium bisulfite conversion of genomic DNA, which is then sequenced on a Next-generation sequencing platform. The sequences obtained are then re-aligned to the reference genome to determine the methylation status of CpG dinucleotides based on mismatches resulting from the conversion of unmethylated cytosines into uracil.
- Enzymatic methyl-seq (EM-seq) works similarly to bisulfite sequencing, but uses enzymes, APOBEC and TET2, to deaminate unmethylated cytosine into uracil prior to sequencing. EM-seq libraries are less prone to DNA damage than bisulfite-treated libraries.[101]
- Reduced representation bisulfite sequencing, also known as RRBS knows several working protocols. The first RRBS protocol was called RRBS and aims for around 10% of the methylome, a reference genome is needed. Later came more protocols that were able to sequence a smaller portion of the genome and higher sample multiplexing. EpiGBS was the first protocol where you could multiplex 96 samples in one lane of Illumina sequencing and were a reference genome was no longer needed. A de novo reference construction from the Watson and Crick reads made population screening of SNP's and SMP's simultaneously a fact.
- The HELP assay, which is based on restriction enzymes' differential ability to recognize and cleave methylated and unmethylated CpG DNA sites.
- GLAD-PCR assay, which is based on a new type of enzymes – site-specific methyl-directed DNA endonucleases, which hydrolyze only methylated DNA.
- ChIP-on-chip assays, which is based on the ability of commercially prepared antibodies to bind to DNA methylation-associated proteins like MeCP2.
- Restriction landmark genomic scanning, a complicated and now rarely used assay based upon restriction enzymes' differential recognition of methylated and unmethylated CpG sites; the assay is similar in concept to the HELP assay.
- Methylated DNA immunoprecipitation (MeDIP), analogous to chromatin immunoprecipitation, immunoprecipitation is used to isolate methylated DNA fragments for input into DNA detection methods such as DNA microarrays (MeDIP-chip) or DNA sequencing (MeDIP-seq).
- Pyrosequencing of bisulfite treated DNA. This is the sequencing of an amplicon made by a normal forward primer but a biotinylated reverse primer to PCR the gene of choice. The Pyrosequencer then analyses the sample by denaturing the DNA and adding one nucleotide at a time to the mix according to a sequence given by the user. If there is a mismatch, it is recorded and the percentage of DNA for which the mismatch is present is noted. This gives the user a percentage of methylation per CpG island.
- Molecular break light assay for DNA adenine methyltransferase activity – an assay that relies on the specificity of the restriction enzyme DpnI for fully methylated (adenine methylation) GATC sites in an oligonucleotide labeled with a fluorophore and quencher. The adenine methyltransferase methylates the oligonucleotide making it a substrate for DpnI. Cutting of the oligonucleotide by DpnI gives rise to a fluorescence increase.[102][103]
- Methyl Sensitive Southern Blotting is similar to the HELP assay, although uses Southern blotting techniques to probe gene-specific differences in methylation using restriction digests. This technique is used to evaluate local methylation near the binding site for the probe.
- MethylCpG Binding Proteins (MBPs) and fusion proteins containing just the Methyl Binding Domain (MBD) are used to separate native DNA into methylated and unmethylated fractions. The percentage methylation of individual CpG islands can be determined by quantifying the amount of the target in each fraction.[citation needed] Extremely sensitive detection can be achieved in FFPE tissues with abscription-based detection.
- High Resolution Melt Analysis (HRM or HRMA), is a post-PCR analytical technique. The target DNA is treated with sodium bisulfite, which chemically converts unmethylated cytosines into uracils, while methylated cytosines are preserved. PCR amplification is then carried out with primers designed to amplify both methylated and unmethylated templates. After this amplification, highly methylated DNA sequences contain a higher number of CpG sites compared to unmethylated templates, which results in a different melting temperature that can be used in quantitative methylation detection.[104][105]
- Ancient DNA methylation reconstruction, a method to reconstruct high-resolution DNA methylation from ancient DNA samples. The method is based on the natural degradation processes that occur in ancient DNA: with time, methylated cytosines are degraded into thymines, whereas unmethylated cytosines are degraded into uracils. This asymmetry in degradation signals was used to reconstruct the full methylation maps of the Neanderthal and the Denisovan.[106] In September 2019, researchers published a novel method to infer morphological traits from DNA methylation data. The authors were able to show that linking down-regulated genes to phenotypes of monogenic diseases, where one or two copies of a gene are perturbed, allows for ~85% accuracy in reconstructing anatomical traits directly from DNA methylation maps.[107]
- Methylation Sensitive Single Nucleotide Primer Extension Assay (msSNuPE), which uses internal primers annealing straight 5' of the nucleotide to be detected.[108]
- Illumina Methylation Assay measures locus-specific DNA methylation using array hybridization. Bisulfite-treated DNA is hybridized to probes on "BeadChips." Single-base base extension with labeled probes is used to determine methylation status of target sites.[109] In 2016, the Infinium MethylationEPIC BeadChip was released, which interrogates over 850,000 methylation sites across the human genome.[110]
- Using nanopore sequencing, researchers have directly identified DNA and RNA base modifications at nucleotide resolution, including 5mC, 5hmC, 6mA, and BrdU in DNA, and m6A in RNA, with detection of other natural or synthetic epigenetic modifications possible through training basecalling algorithms.[111]
Differentially methylated regions (DMRs)[edit]
Differentially methylated regions, which are genomic regions with different methylation statuses among multiple samples (tissues, cells, individuals or others), are regarded as possible functional regions involved in gene transcriptional regulation. The identification of DMRs among multiple tissues (T-DMRs) provides a comprehensive survey of epigenetic differences among human tissues.[112] For example, these methylated regions that are unique to a particular tissue allow individuals to differentiate between tissue type, such as semen and vaginal fluid. Current research conducted by Lee et al., showed DACT1 and USP49 positively identified semen by examining T-DMRs.[113] The use of T-DMRs has proven useful in the identification of various body fluids found at crime scenes. Researchers in the forensic field are currently seeking novel T-DMRs in genes to use as markers in forensic DNA analysis. DMRs between cancer and normal samples (C-DMRs) demonstrate the aberrant methylation in cancers.[114] It is well known that DNA methylation is associated with cell differentiation and proliferation.[115] Many DMRs have been found in the development stages (D-DMRs)[116] and in the reprogrammed progress (R-DMRs).[117] In addition, there are intra-individual DMRs (Intra-DMRs) with longitudinal changes in global DNA methylation along with the increase of age in a given individual.[118] There are also inter-individual DMRs (Inter-DMRs) with different methylation patterns among multiple individuals.[119]
QDMR (Quantitative Differentially Methylated Regions) is a quantitative approach to quantify methylation difference and identify DMRs from genome-wide methylation profiles by adapting Shannon entropy.[120] The platform-free and species-free nature of QDMR makes it potentially applicable to various methylation data. This approach provides an effective tool for the high-throughput identification of the functional regions involved in epigenetic regulation. QDMR can be used as an effective tool for the quantification of methylation difference and identification of DMRs across multiple samples.[121]
Gene-set analysis (a.k.a. pathway analysis; usually performed tools such as DAVID, GoSeq or GSEA) has been shown to be severely biased when applied to high-throughput methylation data (e.g. MeDIP-seq, MeDIP-ChIP, HELP-seq etc.), and a wide range of studies have thus mistakenly reported hyper-methylation of genes related to development and differentiation; it has been suggested that this can be corrected using sample label permutations or using a statistical model to control for differences in the numbers of CpG probes / CpG sites that target each gene.[122]
DNA methylation marks[edit]
DNA methylation marks – genomic regions with specific methylation patterns in a specific biological state such as tissue, cell type, individual – are regarded as possible functional regions involved in gene transcriptional regulation. Although various human cell types may have the same genome, these cells have different methylomes. The systematic identification and characterization of methylation marks across cell types are crucial to understanding the complex regulatory network for cell fate determination. Hongbo Liu et al. proposed an entropy-based framework termed SMART to integrate the whole genome bisulfite sequencing methylomes across 42 human tissues/cells and identified 757,887 genome segments.[123] Nearly 75% of the segments showed uniform methylation across all cell types. From the remaining 25% of the segments, they identified cell type-specific hypo/hypermethylation marks that were specifically hypo/hypermethylated in a minority of cell types using a statistical approach and presented an atlas of the human methylation marks. Further analysis revealed that the cell type-specific hypomethylation marks were enriched through H3K27ac and transcription factor binding sites in a cell type-specific manner. In particular, they observed that the cell type-specific hypomethylation marks are associated with the cell type-specific super-enhancers that drive the expression of cell identity genes. This framework provides a complementary, functional annotation of the human genome and helps to elucidate the critical features and functions of cell type-specific hypomethylation.[citation needed]
The entropy-based Specific Methylation Analysis and Report Tool, termed "SMART", which focuses on integrating a large number of DNA methylomes for the de novo identification of cell type-specific methylation marks. The latest version of SMART is focused on three main functions including de novo identification of differentially methylated regions (DMRs) by genome segmentation, identification of DMRs from predefined regions of interest, and identification of differentially methylated CpG sites.[124]
In identification and detection of body fluids[edit]
DNA methylation allows for several tissues to be analyzed in one assay as well as for small amounts of body fluid to be identified with the use of extracted DNA. Usually, the two approaches of DNA methylation are either methylated-sensitive restriction enzymes or treatment with sodium bisulphite.[125] Methylated sensitive restriction enzymes work by cleaving specific CpG, cytosine and guanine separated by only one phosphate group, recognition sites when the CpG is methylated. In contrast, unmethylated cytosines are transformed to uracil and in the process, methylated cytosines remain methylated. In particular, methylation profiles can provide insight on when or how body fluids were left at crime scenes, identify the kind of body fluid, and approximate age, gender, and phenotypic characteristics of perpetrators.[126] Research indicates various markers that can be used for DNA methylation. Deciding which marker to use for an assay is one of the first steps of the identification of body fluids. In general, markers are selected by examining prior research conducted. Identification markers that are chosen should give a positive result for one type of cell. One portion of the chromosome that is an area of focus when conducting DNA methylation are tissue-specific differentially methylated regions, T-DMRs. The degree of methylation for the T-DMRs ranges depending on the body fluid.[126] A research team developed a marker system that is two-fold. The first marker is methylated only in the target fluid while the second is methylated in the rest of the fluids.[108] For instance, if venous blood marker A is un-methylated and venous blood marker B is methylated in a fluid, it indicates the presence of only venous blood. In contrast, if venous blood marker A is methylated and venous blood marker B is un-methylated in some fluid, then that indicates venous blood is in a mixture of fluids. Some examples for DNA methylation markers are Mens1(menstrual blood), Spei1(saliva), and Sperm2(seminal fluid).
DNA methylation provides a relatively good means of sensitivity when identifying and detecting body fluids. In one study, only ten nanograms of a sample was necessary to ascertain successful results.[127] DNA methylation provides a good discernment of mixed samples since it involves markers that give "on or off" signals. DNA methylation is not impervious to external conditions. Even under degraded conditions using the DNA methylation techniques, the markers are stable enough that there are still noticeable differences between degraded samples and control samples. Specifically, in one study, it was found that there were not any noticeable changes in methylation patterns over an extensive period of time.[126]
The detection of DNA methylation in cell-free DNA and other body fluids has recently become one of the main approaches to Liquid biopsy.[128] In particular, the identification of tissue-specific and disease-specific patterns allows for non-invasive detection and monitoring of diseases such as cancer.[129] If compared to strictly genomic approaches to liquid biopsy, DNA methylation profiling offers a larger number of differentially methylated CpG sites and differentially methylated regions (DMRSs), potentially enhancing its sensitivity. Signal deconvolution algorithms based on DNA methylation have been successfully applied to cell-free DNA and can nominate the tissue of origin of cancers of unknown primary, allograft rejection, and resistance to hormone therapy.[130]
Computational prediction[edit]
DNA methylation can also be detected by computational models through sophisticated algorithms and methods. Computational models can facilitate the global profiling of DNA methylation across chromosomes, and often such models are faster and cheaper to perform than biological assays. Such up-to-date computational models include Bhasin, et al.,[131] Bock, et al.,[132] and Zheng, et al.[133][134] Together with biological assay, these methods greatly facilitate the DNA methylation analysis.
See also[edit]
- 5-Hydroxymethylcytosine
- 5-Methylcytosine
- 7-Methylguanosine
- Decrease in DNA Methylation I (DDM1), a plant methylation gene
- Demethylating agent
- Differentially methylated regions
- DNA demethylation
- DNA methylation reprogramming
- Epigenetics, of which DNA methylation is a significant contributor
- Epigenetic clock, a method to calculate age based on DNA methylation
- Epigenome
- Genome
- Genomic imprinting, an inherited repression of an allele, relying on DNA methylation
- MethBase DNA Methylation database hosted on the UCSC Genome Browser
- MethDB DNA Methylation database
- N6-Methyladenosine
References[edit]
- ^ D. B. Dunn, J. D. Smith: "The occurrence of 6-methylaminopurine in deoxyribonucleic acids". In: Biochem J. 68(4), Apr 1958, S. 627–636. PMID 13522672. PMC 1200409.
- ^ B. F. Vanyushin, S. G. Tkacheva, A. N. Belozersky: "Rare bases in animal DNA". In: Nature. 225, 1970, S. 948–949. PMID 4391887.
- ^ Melanie Ehrlich, Miguel A. Gama-Sosa, Laura H. Carreira, Lars G. Ljungdahl, Kenneth C. Kuo, Charles W. Gehrke: "DNA methylation in thermophilic bacteria: N6-methylcytosine, 5-methylcytosine, and N6-methyladenine." In: Nucleic Acids Research. 13, 1985, S. 1399. PMID 4000939. PMC 341080.
- ^ a b Evans HH, Evans TE (December 1970). "Methylation of the deoxyribonucleic acid of Physarum polycephalum at various periods during the mitotic cycle". The Journal of Biological Chemistry. 245 (23): 6436–6441. doi:10.1016/S0021-9258(18)62627-4. PMID 5530731.

- ^ Smith SS, Rather DI (July 1991). "Lack of 5-methylcytosine in Dictyostelium discoideum DNA". The Biochemical Journal. 277 (1): 273–275. doi:10.1042/bj2770273. PMC 1151219. PMID 1713034.
- ^ a b Drewell RA, Cormier TC, Steenwyk JL, St Denis J, Tabima JF, Dresch JM, Larochelle DA (April 2023). "The Dictyostelium discoideum genome lacks significant DNA methylation and uncovers palindromic sequences as a source of false positives in bisulfite sequencing". NAR genomics bioinformatics. 5 (2): lqad035. doi:10.1093/nargab/lqad035. PMID 37081864.
- ^ Hu CW, Chen JL, Hsu YW, Yen CC, Chao MR (January 2015). "Trace analysis of methylated and hydroxymethylated cytosines in DNA by isotope-dilution LC-MS/MS: first evidence of DNA methylation in Caenorhabditis elegans". The Biochemical Journal. 465 (1): 39–47. doi:10.1042/bj20140844. PMID 25299492.
- ^ Bird A (December 2001). "Molecular biology. Methylation talk between histones and DNA". Science's Compass. Science. 294 (5549): 2113–2115. doi:10.1126/science.1066726. hdl:1842/464. PMID 11739943. S2CID 82947750.
As a result of this process, known as repeat-induced point mutation (RIP), the wild-type Neurospora genome contains a small fraction of methylated DNA, the majority of the DNA remaining nonmethylated.

- ^ a b c Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M (April 2014). "Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species". Analytical Chemistry. 86 (8): 3697–3702. doi:10.1021/ac500447w. PMC 4006885. PMID 24640988.
- ^ Ratel D, Ravanat JL, Berger F, Wion D (March 2006). "N6-methyladenine: the other methylated base of DNA". BioEssays. 28 (3): 309–315. doi:10.1002/bies.20342. PMC 2754416. PMID 16479578.
- ^ Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. (April 2016). "DNA methylation on N(6)-adenine in mammalian embryonic stem cells". Nature. 532 (7599): 329–333. Bibcode:2016Natur.532..329W. doi:10.1038/nature17640. PMC 4977844. PMID 27027282.
- ^ Angéla Békési and Beáta G Vértessy "Uracil in DNA: error or signal?"
- ^ Rana AK, Ankri S (2016). "Reviving the RNA World: An Insight into the Appearance of RNA Methyltransferases". Frontiers in Genetics. 7: 99. doi:10.3389/fgene.2016.00099. PMC 4893491. PMID 27375676.
- ^ Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E (May 2002). "De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation". Gene. 289 (1–2): 41–48. doi:10.1016/S0378-1119(02)00469-9. PMID 12036582.
- ^ Haines TR, Rodenhiser DI, Ainsworth PJ (December 2001). "Allele-specific non-CpG methylation of the Nf1 gene during early mouse development". Developmental Biology. 240 (2): 585–598. doi:10.1006/dbio.2001.0504. PMID 11784085.
- ^ a b Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. (November 2009). "Human DNA methylomes at base resolution show widespread epigenomic differences". Nature. 462 (7271): 315–322. Bibcode:2009Natur.462..315L. doi:10.1038/nature08514. PMC 2857523. PMID 19829295.
- ^ Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. (August 2013). "Global epigenomic reconfiguration during mammalian brain development". Science. 341 (6146): 1237905. doi:10.1126/science.1237905. PMC 3785061. PMID 23828890.
- ^ a b Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G, et al. (July 2015). "Whole-genome fingerprint of the DNA methylome during human B cell differentiation". Nature Genetics. 47 (7): 746–756. doi:10.1038/ng.3291. PMC 5444519. PMID 26053498.
- ^ Tost J (January 2010). "DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker". Molecular Biotechnology. 44 (1): 71–81. doi:10.1007/s12033-009-9216-2. PMID 19842073. S2CID 20307488.
- ^ Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. (December 2011). "DNA-binding factors shape the mouse methylome at distal regulatory regions". Nature. 480 (7378): 490–495. doi:10.1038/nature11086. PMID 22170606.
- ^ a b c Zemach A, McDaniel IE, Silva P, Zilberman D (May 2010). "Genome-wide evolutionary analysis of eukaryotic DNA methylation". Science (ScienceExpress Report). 328 (5980): 916–919. Bibcode:2010Sci...328..916Z. doi:10.1126/science.1186366. PMID 20395474. S2CID 206525166.
Here we quantify DNA methylation in seventeen eukaryotic genomes....
 Supplemental figures appear to be only accessible via the science.sciencemag.org paywall.
Supplemental figures appear to be only accessible via the science.sciencemag.org paywall.
- ^ Suzuki MM, Kerr AR, De Sousa D, Bird A (May 2007). "CpG methylation is targeted to transcription units in an invertebrate genome". Genome Research. 17 (5): 625–631. doi:10.1101/gr.6163007. PMC 1855171. PMID 17420183.
- ^ a b Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. (February 2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. Bibcode:2001Natur.409..860L. doi:10.1038/35057062. hdl:2027.42/62798. PMID 11237011.
- ^ Bird AP (1986-05-15). "CpG-rich islands and the function of DNA methylation". Nature. 321 (6067): 209–213. Bibcode:1986Natur.321..209B. doi:10.1038/321209a0. PMID 2423876. S2CID 4236677.
- ^ Gardiner-Garden M, Frommer M (July 1987). "CpG islands in vertebrate genomes". Journal of Molecular Biology. 196 (2): 261–282. doi:10.1016/0022-2836(87)90689-9. PMID 3656447.
- ^ Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, et al. (September 2010). "Orphan CpG islands identify numerous conserved promoters in the mammalian genome". PLOS Genetics. 6 (9): e1001134. doi:10.1371/journal.pgen.1001134. PMC 2944787. PMID 20885785.
- ^ Saxonov S, Berg P, Brutlag DL (January 2006). "A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters". Proceedings of the National Academy of Sciences of the United States of America. 103 (5): 1412–1417. Bibcode:2006PNAS..103.1412S. doi:10.1073/pnas.0510310103. PMC 1345710. PMID 16432200.
- ^ a b Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, et al. (May 2010). "Conservation and divergence of methylation patterning in plants and animals". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8689–8694. doi:10.1073/pnas.1002720107. PMC 2889301. PMID 20395551.
- ^ Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, et al. (June 2008). "Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors". Molecular Cell. 30 (6): 755–766. doi:10.1016/j.molcel.2008.05.007. PMID 18514006.
- ^ Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D (April 2007). "Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome". Nature Genetics. 39 (4): 457–466. doi:10.1038/ng1990. PMID 17334365. S2CID 22446734.
- ^ Schübeler D (January 2015). "Function and information content of DNA methylation". Nature. 517 (7534): 321–326. Bibcode:2015Natur.517..321S. doi:10.1038/nature14192. PMID 25592537. S2CID 4403755.
- ^ Choy MK, Movassagh M, Goh HG, Bennett MR, Down TA, Foo RS (September 2010). "Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated". BMC Genomics. 11 (1): 519. doi:10.1186/1471-2164-11-519. PMC 2997012. PMID 20875111.
- ^ Dahlet T, Argüeso Lleida A, Al Adhami H, Dumas M, Bender A, Ngondo RP, et al. (June 2020). "Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity". Nature Communications. 11 (1): 3153. Bibcode:2020NatCo..11.3153D. doi:10.1038/s41467-020-16919-w. PMC 7305168. PMID 32561758.
- ^ Huff JT, Zilberman D (March 2014). "Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes". Cell. 156 (6): 1286–1297. doi:10.1016/j.cell.2014.01.029. PMC 3969382. PMID 24630728.
- ^ Yoder JA, Walsh CP, Bestor TH (August 1997). "Cytosine methylation and the ecology of intragenomic parasites". Trends in Genetics. 13 (8): 335–340. doi:10.1016/s0168-9525(97)01181-5. PMID 9260521.
- ^ Zhou W, Liang G, Molloy PL, Jones PA (August 2020). "DNA methylation enables transposable element-driven genome expansion". Proceedings of the National Academy of Sciences of the United States of America. 117 (32): 19359–19366. Bibcode:2020PNAS..11719359Z. doi:10.1073/pnas.1921719117. PMC 7431005. PMID 32719115.
- ^ Lev Maor G, Yearim A, Ast G (May 2015). "The alternative role of DNA methylation in splicing regulation". Trends in Genetics. 31 (5): 274–280. doi:10.1016/j.tig.2015.03.002. PMID 25837375. S2CID 34258335.
- ^ Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. (July 2010). "Conserved role of intragenic DNA methylation in regulating alternative promoters". Nature. 466 (7303): 253–257. Bibcode:2010Natur.466..253M. doi:10.1038/nature09165. PMC 3998662. PMID 20613842.
- ^ Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. (November 2005). "Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription". Cell. 123 (4): 581–592. doi:10.1016/j.cell.2005.10.023. PMID 16286007. S2CID 9328002.
- ^ Cedar H, Bergman Y (July 2012). "Programming of DNA methylation patterns". Annual Review of Biochemistry. 81: 97–117. doi:10.1146/annurev-biochem-052610-091920. PMID 22404632. – via Annual Reviews (subscription required)
- ^ a b Mužić Radović V, Bunoza P, Marić T, Himelreich-Perić M, Bulić-Jakuš F, Takahashi M, et al. (July 2022). "Global DNA methylation and chondrogenesis of rat limb buds in a three-dimensional organ culture system". Bosnian Journal of Basic Medical Sciences. 22 (4): 560–568. doi:10.17305/bjbms.2021.6584. PMC 9392980. PMID 35188093. S2CID 247010798.
- ^ Beard C, Li E, Jaenisch R (October 1995). "Loss of methylation activates Xist in somatic but not in embryonic cells". Genes & Development. 9 (19): 2325–2334. doi:10.1101/gad.9.19.2325. PMID 7557385.
- ^ Li E, Beard C, Jaenisch R (November 1993). "Role for DNA methylation in genomic imprinting". Nature. 366 (6453): 362–365. Bibcode:1993Natur.366..362L. doi:10.1038/366362a0. PMID 8247133. S2CID 4311091.
- ^ Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, et al. (December 2010). "Targets and dynamics of promoter DNA methylation during early mouse development". Nature Genetics. 42 (12): 1093–1100. doi:10.1038/ng.708. PMID 21057502. S2CID 205357042.
- ^ Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W (January 2013). "Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 368 (1609): 20110330. doi:10.1098/rstb.2011.0330. PMC 3539359. PMID 23166394.
- ^ Wang YP, Lei QY (May 2018). "Metabolic recoding of epigenetics in cancer". Cancer Communications. 38 (1): 25. doi:10.1186/s40880-018-0302-3. PMC 5993135. PMID 29784032.
- ^ Daura-Oller E, Cabre M, Montero MA, Paternain JL, Romeu A (April 2009). "Specific gene hypomethylation and cancer: new insights into coding region feature trends". Bioinformation. 3 (8): 340–343. doi:10.6026/97320630003340. PMC 2720671. PMID 19707296.
- ^ Craig JM, Wong NC, eds. (2011). Epigenetics: A Reference Manual. Caister Academic Press. ISBN 978-1-904455-88-2.
- ^ a b c Gonzalo S (August 2010). "Epigenetic alterations in aging". Journal of Applied Physiology. 109 (2): 586–597. doi:10.1152/japplphysiol.00238.2010. PMC 2928596. PMID 20448029.
- ^ Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, et al. (July 2004). "DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E". The Journal of Biological Chemistry. 279 (28): 29147–29154. doi:10.1074/jbc.m403618200. hdl:2445/176763. PMID 15131116.
- ^ Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, et al. (August 2003). "Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease". Clinical Chemistry. 49 (8): 1292–1296. doi:10.1373/49.8.1292. PMID 12881445.
- ^ Huang YS, Zhi YF, Wang SR (October 2009). "Hypermethylation of estrogen receptor-alpha gene in atheromatosis patients and its correlation with homocysteine". Pathophysiology. 16 (4): 259–265. doi:10.1016/j.pathophys.2009.02.010. PMID 19285843.
- ^ Dong C, Yoon W, Goldschmidt-Clermont PJ (August 2002). "DNA methylation and atherosclerosis". The Journal of Nutrition. 132 (8 Suppl): 2406S–2409S. doi:10.1093/jn/132.8.2406S. PMID 12163701.
- ^ Ying AK, Hassanain HH, Roos CM, Smiraglia DJ, Issa JJ, Michler RE, et al. (April 2000). "Methylation of the estrogen receptor-alpha gene promoter is selectively increased in proliferating human aortic smooth muscle cells". Cardiovascular Research. 46 (1): 172–179. doi:10.1016/s0008-6363(00)00004-3. PMID 10727665.
- ^ Zhu S, Goldschmidt-Clermont PJ, Dong C (August 2005). "Inactivation of monocarboxylate transporter MCT3 by DNA methylation in atherosclerosis". Circulation. 112 (9): 1353–1361. doi:10.1161/circulationaha.104.519025. PMID 16116050.
- ^ Pepin ME, Ha CM, Crossman DK, Litovsky SH, Varambally S, Barchue JP, et al. (March 2019). "Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure". Laboratory Investigation; A Journal of Technical Methods and Pathology. 99 (3): 371–386. doi:10.1038/s41374-018-0104-x. PMC 6515060. PMID 30089854.
- ^ Pepin ME, Ha CM, Potter LA, Bakshi S, Barchue JP, Haj Asaad A, et al. (May 2021). "Racial and socioeconomic disparity associates with differences in cardiac DNA methylation among men with end-stage heart failure". American Journal of Physiology. Heart and Circulatory Physiology. 320 (5): H2066–H2079. doi:10.1152/ajpheart.00036.2021. PMC 8163657. PMID 33769919.
- ^ Horvath S (2013). "DNA methylation age of human tissues and cell types". Genome Biology. 14 (10): R115. doi:10.1186/gb-2013-14-10-r115. PMC 4015143. PMID 24138928.
- ^ Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, et al. (August 2010). "A longitudinal study of epigenetic variation in twins". Epigenetics. 5 (6): 516–526. doi:10.4161/epi.5.6.12226. PMC 3322496. PMID 20505345.
- ^ a b Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, et al. (June 2012). "Distinct DNA methylomes of newborns and centenarians". Proceedings of the National Academy of Sciences of the United States of America. 109 (26): 10522–10527. Bibcode:2012PNAS..10910522H. doi:10.1073/pnas.1120658109. PMC 3387108. PMID 22689993.
- ^ a b Barrès R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. (March 2012). "Acute exercise remodels promoter methylation in human skeletal muscle". Cell Metabolism. 15 (3): 405–411. doi:10.1016/j.cmet.2012.01.001. PMID 22405075.
- ^ Rönn T, Volkov P, Davegårdh C, Dayeh T, Hall E, Olsson AH, et al. (June 2013). "A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue". PLOS Genetics. 9 (6): e1003572. doi:10.1371/journal.pgen.1003572. PMC 3694844. PMID 23825961.
- ^ Zhang FF, Cardarelli R, Carroll J, Zhang S, Fulda KG, Gonzalez K, et al. (March 2011). "Physical activity and global genomic DNA methylation in a cancer-free population". Epigenetics. 6 (3): 293–299. doi:10.4161/epi.6.3.14378. PMC 3092677. PMID 21178401.
- ^ a b Sweatt JD (May 2016). "Dynamic DNA methylation controls glutamate receptor trafficking and synaptic scaling". Journal of Neurochemistry. 137 (3): 312–330. doi:10.1111/jnc.13564. PMC 4836967. PMID 26849493.
- ^ Kim S, Kaang BK (January 2017). "Epigenetic regulation and chromatin remodeling in learning and memory". Experimental & Molecular Medicine. 49 (1): e281. doi:10.1038/emm.2016.140. PMC 5291841. PMID 28082740.
- ^ Schafe GE, Nadel NV, Sullivan GM, Harris A, LeDoux JE (1999). "Memory consolidation for contextual and auditory fear conditioning is dependent on protein synthesis, PKA, and MAP kinase". Learning & Memory. 6 (2): 97–110. doi:10.1101/lm.6.2.97. PMC 311283. PMID 10327235.
- ^ a b c Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, et al. (January 2016). "DNA methylation changes in plasticity genes accompany the formation and maintenance of memory". Nature Neuroscience. 19 (1): 102–110. doi:10.1038/nn.4194. PMC 4700510. PMID 26656643.
- ^ a b Duke CG, Kennedy AJ, Gavin CF, Day JJ, Sweatt JD (July 2017). "Experience-dependent epigenomic reorganization in the hippocampus". Learning & Memory. 24 (7): 278–288. doi:10.1101/lm.045112.117. PMC 5473107. PMID 28620075.
- ^ a b Bernstein C (2022). "DNA Methylation and Establishing Memory". Epigenetics Insights. 15: 25168657211072499. doi:10.1177/25168657211072499. PMC 8793415. PMID 35098021.
- ^ Gratchev A. "Review on DNA Methylation". METHODS.info.
- ^ Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, et al. (November 2016). "The DNA methyltransferase DNMT3C protects male germ cells from transposon activity". Science. 354 (6314): 909–912. Bibcode:2016Sci...354..909B. doi:10.1126/science.aah5143. PMID 27856912. S2CID 30907442.
- ^ Jain D, Meydan C, Lange J, Claeys Bouuaert C, Lailler N, Mason CE, et al. (August 2017). "rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline". PLOS Genetics. 13 (8): e1006964. doi:10.1371/journal.pgen.1006964. PMC 5607212. PMID 28854222.
- ^ Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. (January 2006). "Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2". Science. 311 (5759): 395–398. Bibcode:2006Sci...311..395G. doi:10.1126/science.1120976. PMID 16424344. S2CID 39089541.
- ^ a b Zhang H, Lang Z, Zhu JK (August 2018). "Dynamics and function of DNA methylation in plants". Nature Reviews. Molecular Cell Biology. 19 (8): 489–506. doi:10.1038/s41580-018-0016-z. PMID 29784956. S2CID 256745172.
- ^ Dubin MJ, Zhang P, Meng D, Remigereau MS, Osborne EJ, Paolo Casale F, et al. (May 2015). Dermitzakis ET (ed.). "DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation". eLife. 4: e05255. doi:10.7554/eLife.05255. PMC 4413256. PMID 25939354.
- ^ Cao X, Jacobsen SE (December 2002). "Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes". Proceedings of the National Academy of Sciences of the United States of America. 99 (Suppl 4): 16491–16498. Bibcode:2002PNAS...9916491C. doi:10.1073/pnas.162371599. PMC 139913. PMID 12151602.
- ^ Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, et al. (September 2016). "Widespread natural variation of DNA methylation within angiosperms". Genome Biology. 17 (1): 194. doi:10.1186/s13059-016-1059-0. PMC 5037628. PMID 27671052.
- ^ Zemach A, McDaniel IE, Silva P, Zilberman D (May 2010). "Genome-wide evolutionary analysis of eukaryotic DNA methylation". Science. 328 (5980): 916–919. Bibcode:2010Sci...328..916Z. doi:10.1126/science.1186366. PMID 20395474. S2CID 206525166.
- ^ a b Aufsatz W, Mette MF, van der Winden J, Matzke AJ, Matzke M (December 2002). "RNA-directed DNA methylation in Arabidopsis". Proceedings of the National Academy of Sciences of the United States of America. 99 (Suppl 4): 16499–16506. Bibcode:2002PNAS...9916499A. doi:10.1073/pnas.162371499. PMC 139914. PMID 12169664.
- ^ Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ (March 2017). "Evolution of DNA Methylation across Insects". Molecular Biology and Evolution. 34 (3): 654–665. doi:10.1093/molbev/msw264. PMC 5400375. PMID 28025279.
- ^ Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, et al. (October 2006). "Functional CpG methylation system in a social insect". Science. 314 (5799): 645–647. Bibcode:2006Sci...314..645W. doi:10.1126/science.1135213. PMID 17068262. S2CID 31709665.
- ^ Ying and Li-Byarlay (2015). Physiological and Molecular Mechanisms of Nutrition in Honey Bees. Advances in Insect Physiology. Vol. 49. pp. 25–58. doi:10.1016/bs.aiip.2015.06.002. ISBN 9780128025864.
- ^ Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, Kaneda M, et al. (July 2013). "RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee". Proceedings of the National Academy of Sciences of the United States of America. 110 (31): 12750–12755. Bibcode:2013PNAS..11012750L. doi:10.1073/pnas.1310735110. PMC 3732956. PMID 23852726.
- ^ Smith SS, Thomas CA (May 1981). "The two-dimensional restriction analysis of Drosophila DNAs: males and females". Gene. 13 (4): 395–408. doi:10.1016/0378-1119(81)90019-6. PMID 6266924.
- ^ Lyko F, Ramsahoye BH, Jaenisch R (November 2000). "DNA methylation in Drosophila melanogaster". Nature. 408 (6812): 538–540. doi:10.1038/35046205. PMID 11117732. S2CID 4427540.
- ^ Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, et al. (May 2014). "Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity". Genome Research. 24 (5): 821–830. doi:10.1101/gr.162412.113. PMC 4009611. PMID 24558263.
- ^ Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, et al. (May 2015). "N6-methyladenine DNA modification in Drosophila". Cell. 161 (4): 893–906. doi:10.1016/j.cell.2015.04.018. PMID 25936838.
- ^ Antequera F, Tamame M, Villanueva JR, Santos T (July 1984). "DNA methylation in the fungi". The Journal of Biological Chemistry. 259 (13): 8033–8036. doi:10.1016/S0021-9258(17)39681-3. PMID 6330093.
- ^ Binz T, D'Mello N, Horgen PA (1998). "A comparison of DNA methylation levels in selected isolates of higher fungi". Mycologia. 90 (5): 785–790. doi:10.2307/3761319. JSTOR 3761319.
- ^ Liu SY, Lin JQ, Wu HL, Wang CC, Huang SJ, Luo YF, et al. (2012). "Bisulfite sequencing reveals that Aspergillus flavus holds a hollow in DNA methylation". PLOS ONE. 7 (1): e30349. Bibcode:2012PLoSO...730349L. doi:10.1371/journal.pone.0030349. PMC 3262820. PMID 22276181.
- ^ Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M (April 2003). "The methylated component of the Neurospora crassa genome". Nature. 422 (6934): 893–897. Bibcode:2003Natur.422..893S. doi:10.1038/nature01564. hdl:1842/694. PMID 12712205. S2CID 4380222.
- ^ Smith SS, Ratner DI (July 1991). "Lack of 5-methylcytosine in Dictyostelium discoideum DNA". The Biochemical Journal. 277 ( Pt 1) (Pt 1): 273–275. doi:10.1042/bj2770273. PMC 1151219. PMID 1713034.
- ^ Reilly JG, Braun R, Thomas CA (July 1980). "Methjylation in Physarum DNA". FEBS Letters. 116 (2): 181–184. doi:10.1016/0014-5793(80)80638-7. PMID 6250882. S2CID 83941623.
- ^ Matthew J. Blow, Tyson A. Clark, Chris G. Daum, Adam M. Deutschbauer, Alexey Fomenkov, Roxanne Fries, Jeff Froula, Dongwan D. Kang, Rex R. Malmstrom, Richard D. Morgan, Janos Posfai, Kanwar Singh, Axel Visel, Kelly Wetmore, Zhiying Zhao, Edward M. Rubin, Jonas Korlach, Len A. Pennacchio, Richard J. Roberts: The Epigenomic Landscape of Prokaryotes. In: PLoS Genet. 12(2), Feb 2016, S. e1005854. doi:10.1371/journal.pgen.1005854. PMID 26870957. PMC 4752239
- ^ Oliveira PH (August 2021). "Bacterial Epigenomics: Coming of Age". mSystems. 6 (4): e0074721. doi:10.1128/mSystems.00747-21. PMC 8407109. PMID 34402642.
- ^ Oliveira PH, Ribis JW, Garrett EM, Trzilova D, Kim A, Sekulovic O, et al. (January 2020). "Epigenomic characterization of Clostridioides difficile finds a conserved DNA methyltransferase that mediates sporulation and pathogenesis". Nature Microbiology. 5 (1): 166–180. doi:10.1038/s41564-019-0613-4. PMC 6925328. PMID 31768029.
- ^ Palmer BR, Marinus MG (May 1994). "The dam and dcm strains of Escherichia coli--a review". Gene. 143 (1): 1–12. doi:10.1016/0378-1119(94)90597-5. PMID 8200522.
- ^ "Making unmethylated (dam-/dcm-) DNA". Archived from the original on 2011-01-06.
- ^ Rana AK (January 2018). "Crime investigation through DNA methylation analysis: methods and applications in forensics". Egyptian Journal of Forensic Sciences. 8 (7). doi:10.1186/s41935-018-0042-1.
- ^ Hernández HG, Tse MY, Pang SC, Arboleda H, Forero DA (October 2013). "Optimizing methodologies for PCR-based DNA methylation analysis". BioTechniques. 55 (4): 181–197. doi:10.2144/000114087. PMID 24107250.
- ^ Feng, Suhua; Zhong, Zhenhui; Wang, Ming; Jacobsen, Steven E. (2020-10-07). "Efficient and accurate determination of genome-wide DNA methylation patterns in Arabidopsis thaliana with enzymatic methyl sequencing". Epigenetics & Chromatin. 13 (1): 42. doi:10.1186/s13072-020-00361-9. ISSN 1756-8935. PMC 7542392. PMID 33028374.
- ^ Wood RJ, Maynard-Smith MD, Robinson VL, Oyston PC, Titball RW, Roach PL (August 2007). Fugmann S (ed.). "Kinetic analysis of Yersinia pestis DNA adenine methyltransferase activity using a hemimethylated molecular break light oligonucleotide". PLOS ONE. 2 (8): e801. Bibcode:2007PLoSO...2..801W. doi:10.1371/journal.pone.0000801. PMC 1949145. PMID 17726531.
- ^ Li J, Yan H, Wang K, Tan W, Zhou X (February 2007). "Hairpin fluorescence DNA probe for real-time monitoring of DNA methylation". Analytical Chemistry. 79 (3): 1050–1056. doi:10.1021/ac061694i. PMID 17263334.
- ^ Wojdacz TK, Dobrovic A (2007). "Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation". Nucleic Acids Research. 35 (6): e41. doi:10.1093/nar/gkm013. PMC 1874596. PMID 17289753.
- ^ Malentacchi F, Forni G, Vinci S, Orlando C (July 2009). "Quantitative evaluation of DNA methylation by optimization of a differential-high resolution melt analysis protocol". Nucleic Acids Research. 37 (12): e86. doi:10.1093/nar/gkp383. PMC 2709587. PMID 19454604.
- ^ Gokhman D, Lavi E, Prüfer K, Fraga MF, Riancho JA, Kelso J, et al. (May 2014). "Reconstructing the DNA methylation maps of the Neandertal and the Denisovan". Science. 344 (6183): 523–527. Bibcode:2014Sci...344..523G. doi:10.1126/science.1250368. PMID 24786081. S2CID 28665590.
- ^ Gokhman D, Mishol N, de Manuel M, de Juan D, Shuqrun J, Meshorer E, et al. (September 2019). "Reconstructing Denisovan Anatomy Using DNA Methylation Maps". Cell. 179 (1): 180–192.e10. doi:10.1016/j.cell.2019.08.035. PMID 31539495. S2CID 202676502.
- ^ a b Forat S, Huettel B, Reinhardt R, Fimmers R, Haidl G, Denschlag D, Olek K (2016-02-01). "Methylation Markers for the Identification of Body Fluids and Tissues from Forensic Trace Evidence". PLOS ONE. 11 (2): e0147973. Bibcode:2016PLoSO..1147973F. doi:10.1371/journal.pone.0147973. PMC 4734623. PMID 26829227.
- ^ "Infinium Methylation Assay | Interrogate single CpG sites". www.illumina.com. Retrieved 2020-01-10.
- ^ "Infinium MethylationEPIC Kit | Methylation profiling array for EWAS". www.illumina.com. Retrieved 2020-01-10.
- ^ "Epigenetics and methylation analysis". Oxford Nanopore Technologies. Archived from the original on 2021-09-01. Retrieved 2021-09-27.
- ^ Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Gräf S, et al. (September 2008). "An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs)". Genome Research. 18 (9): 1518–1529. doi:10.1101/gr.077479.108. PMC 2527707. PMID 18577705.
- ^ Lee HY, Park MJ, Choi A, An JH, Yang WI, Shin KJ (January 2012). "Potential forensic application of DNA methylation profiling to body fluid identification". International Journal of Legal Medicine. 126 (1): 55–62. doi:10.1007/s00414-011-0569-2. PMID 21626087. S2CID 22243051.
- ^ Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. (February 2009). "The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores". Nature Genetics. 41 (2): 178–186. doi:10.1038/ng.298. PMC 2729128. PMID 19151715.
- ^ Reik W, Dean W, Walter J (August 2001). "Epigenetic reprogramming in mammalian development". Science. 293 (5532): 1089–1093. doi:10.1126/science.1063443. PMID 11498579. S2CID 17089710.
- ^ Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. (August 2008). "Genome-scale DNA methylation maps of pluripotent and differentiated cells". Nature. 454 (7205): 766–770. Bibcode:2008Natur.454..766M. doi:10.1038/nature07107. PMC 2896277. PMID 18600261.
- ^ Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. (December 2009). "Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts". Nature Genetics. 41 (12): 1350–1353. doi:10.1038/ng.471. PMC 2958040. PMID 19881528.
- ^ Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. (June 2008). "Intra-individual change over time in DNA methylation with familial clustering". JAMA. 299 (24): 2877–2883. doi:10.1001/jama.299.24.2877. PMC 2581898. PMID 18577732.
- ^ Bock C, Walter J, Paulsen M, Lengauer T (June 2008). "Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping". Nucleic Acids Research. 36 (10): e55. doi:10.1093/nar/gkn122. PMC 2425484. PMID 18413340.
- ^ "QDMR: a quantitative method for identification of differentially methylated regions by entropy". bioinfo.hrbmu.edu.cn. Archived from the original on 2015-10-23. Retrieved 2013-03-09.
- ^ Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, et al. (May 2011). "QDMR: a quantitative method for identification of differentially methylated regions by entropy". Nucleic Acids Research. 39 (9): e58. doi:10.1093/nar/gkr053. PMC 3089487. PMID 21306990.
- ^ Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C (August 2013). "Gene-set analysis is severely biased when applied to genome-wide methylation data". Bioinformatics. 29 (15): 1851–1857. doi:10.1093/bioinformatics/btt311. PMID 23732277.
- ^ Liu H, Liu X, Zhang S, Lv J, Li S, Shang S, et al. (January 2016). "Systematic identification and annotation of human methylation marks based on bisulfite sequencing methylomes reveals distinct roles of cell type-specific hypomethylation in the regulation of cell identity genes". Nucleic Acids Research. 44 (1): 75–94. doi:10.1093/nar/gkv1332. PMC 4705665. PMID 26635396.
- ^ Hongbo L (2016). "SMART 2: A Comprehensive Analysis Tool for Bisulfite Sequencing Data". fame.edbc.org.
- ^ Sijen T (September 2015). "Molecular approaches for forensic cell type identification: On mRNA, miRNA, DNA methylation and microbial markers". Forensic Science International. Genetics. 18: 21–32. doi:10.1016/j.fsigen.2014.11.015. PMID 25488609.
- ^ a b c Kader F, Ghai M (April 2015). "DNA methylation and application in forensic sciences". Forensic Science International. 249: 255–265. doi:10.1016/j.forsciint.2015.01.037. PMID 25732744.
- ^ Silva DS, Antunes J, Balamurugan K, Duncan G, Alho CS, McCord B (July 2016). "Developmental validation studies of epigenetic DNA methylation markers for the detection of blood, semen and saliva samples". Forensic Science International. Genetics. 23: 55–63. doi:10.1016/j.fsigen.2016.01.017. hdl:10923/23633. PMID 27010659. S2CID 7438775.
- ^ Dor Y, Cedar H (September 2018). "Principles of DNA methylation and their implications for biology and medicine". Lancet. 392 (10149): 777–786. doi:10.1016/S0140-6736(18)31268-6. PMID 30100054. S2CID 51965986.
- ^ Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV (June 2020). "Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA". Annals of Oncology. 31 (6): 745–759. doi:10.1016/j.annonc.2020.02.011. PMC 8274402. PMID 33506766.
- ^ Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, et al. (November 2018). "Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease". Nature Communications. 9 (1): 5068. Bibcode:2018NatCo...9.5068M. doi:10.1038/s41467-018-07466-6. PMC 6265251. PMID 30498206.
- ^ Bhasin M, Zhang H, Reinherz EL, Reche PA (August 2005). "Prediction of methylated CpGs in DNA sequences using a support vector machine" (PDF). FEBS Letters. 579 (20): 4302–4308. doi:10.1016/j.febslet.2005.07.002. PMID 16051225. S2CID 14487630.
- ^ Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J (March 2006). "CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure". PLOS Genetics. 2 (3): e26. doi:10.1371/journal.pgen.0020026. PMC 1386721. PMID 16520826.
- ^ Zheng H, Jiang SW, Wu H (2011). "Enhancement on the predictive power of the prediction model for human genomic DNA methylation". International Conference on Bioinformatics and Computational Biology (BIOCOMP'11).
- ^ Zheng H, Wu H, Li J, Jiang SW (2013). "CpGIMethPred: computational model for predicting methylation status of CpG islands in human genome". BMC Medical Genomics. 6 (Suppl 1): S13. doi:10.1186/1755-8794-6-S1-S13. PMC 3552668. PMID 23369266.
Further reading[edit]
- Law JA, Jacobsen SE (March 2010). "Establishing, maintaining and modifying DNA methylation patterns in plants and animals". Nature Reviews. Genetics. 11 (3): 204–220. doi:10.1038/nrg2719. PMC 3034103. PMID 20142834.
- Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, et al. (May 2009). "Developmental programming of CpG island methylation profiles in the human genome". Nature Structural & Molecular Biology. 16 (5): 564–571. doi:10.1038/nsmb.1594. PMID 19377480. S2CID 8804930.
- Patra SK (April 2008). "Ras regulation of DNA-methylation and cancer". Experimental Cell Research. 314 (6): 1193–1201. doi:10.1016/j.yexcr.2008.01.012. PMID 18282569.
- Patra SK, Patra A, Rizzi F, Ghosh TC, Bettuzzi S (June 2008). "Demethylation of (Cytosine-5-C-methyl) DNA and regulation of transcription in the epigenetic pathways of cancer development". Cancer and Metastasis Reviews. 27 (2): 315–334. doi:10.1007/s10555-008-9118-y. hdl:11381/1797001. PMID 18246412. S2CID 22435914.
- Nagpal G, Sharma M, Kumar S, Chaudhary K, Gupta S, Gautam A, Raghava GP (February 2014). "PCMdb: pancreatic cancer methylation database". Scientific Reports. 4: 4197. Bibcode:2014NatSR...4E4197N. doi:10.1038/srep04197. PMC 3935225. PMID 24569397.
External links[edit]
- DNA+Methylation at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- ENCODE threads explorer Non-coding RNA characterization. Nature (journal)
- PCMdb Pancreatic Cancer Methylation Database.
- SMART Specific Methylation Analysis and Report Tool
- Human Methylation Mark Atlas
- DiseaseMeth Archived 2020-01-27 at the Wayback Machine Human disease methylation database
- EWAS Atlas A knowledgebase of epigenome-wide association studies
