User:Williamog1990/sandbox
Cre Recombinase[edit]
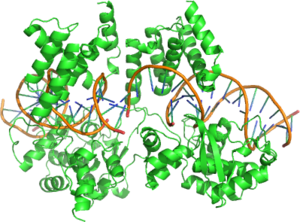
This page is dedicated to the Cre Recombinase Enzyme. For a detailed page on Cre-lox Recombination follow this link: Cre-Lox Recombination
Cre Recombinase is a tyrosine recombinase enzyme derived from the P1 Bacteriophage. The enzyme uses a topoisomerase I like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the Integrase family of site specific recombinase and it is known to catalyse the site specific recombination event between two DNA recognition sites (loxP sites). This 34 base pair (bp) loxP recognition site is comprised of two 13 bp palindromic sequences which flank an 8bp spacer region. The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. DNA sequences found between two loxP sites are said to be "floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites orienated in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are opposingly orientated will be inverted[1]. The enzyme requires no additional cofactors (such as ATP) or accessory proteins for its function[2].
The enzyme plays important roles in the life cycle of the P1 Bacteriophage such as cyclization of the linear genome and resolution of dimeric chromosomes that form after DNA replication[3].
Cre recombinase is a widely used tool in the field of molecular biology. The enzyme’s unique and specific recombination system is exploited to manipulate genes and chromosomes in a huge range of research such as gene knock out or knock in studies. The enzyme’s ability to operate efficiently in a wide range of cellular environments (including mammamals,plants,bacteria and yeast) enables the Cre-Lox Recombination system to be used in a vast number of organisms making it a particularly useful tool in scientific research[4].
Discovery[edit]
Studies carried out in 1981 by Sternberg & Hamilton demonstrated that the bacteriophage P1 had a unique site specific recombination system. EcoRI fragments of the P1 Bacteriophage genome were generated and cloned into lambda vectors. A 6.5x103 base EcoRI fragment (Fragemnt 7) was found to permit efficient recombination events[5]. The mechanism of these recombination events were known to be unique as they occurred in the absence of bacterial RecA and RecBCD proteins. The components of this recombination system were elucidated using deletion mutagenesis studies. These studies showed that a P1gene product and a recombination site were both required for efficient recombination events to occur. The P1 gene product was named Cre(Causes Recombination)and the recombination site was named loxP (locus of crossing (x) over,P1). The Cre protein was purified in 1983 and was found to be a 35,000 Da protein[2]. It was also elucidated at this time that no high energy cofactors such as ATP or accessory proteins are required for the recombinase activity of the purified protein[2]. Early studies also demonstrated that Cre binds to non specific DNA sequences whilst having a 20 fold higher affinity for loxP sequences and results of early DNA footprinting studies also suggested that Cre molecules bind loxP sites as dimers[2].
| Tyrosine Recombinase Family Members[3] |
|---|
| S.cerevisiae Flp Recombinase |
| Bacterial XerC Recombinase |
| Bacterial XerD Recombinase |
| λ Integrase Protein |
| HP1 Integrase Protein |
Structure[edit]
Cre Recombinase is comprised of 343 amino acids that form two distinct domains. The amino terminal domain encompasses residues 20-129 and this domain contains 5 alpha helical segments linked by a series of short loops. Helices A & E are involved in the formation of the recombinase tetramer with the C terminus region of helix E known to form contacts with the C terminal domain of adjacent subunits. Helices B & D form direct contacts with the major groove of the loxP DNA. These two helices are thought to make three direct contacts to DNA bases at the loxP site. The carboxy terminal domain of the enzyme consisits of amino acids 132-341 and it harbours the active site of the enzyme. The overall structure of this domain shares a great deal of structural resemblance to the catalycic domain of other enzymes of the same family such as λ Integrase and HP1 Integrase. This domain is predominantly helical in structure with 9 distinct helices (F-N). The terminal helix (N) protrudes from the main body of the carboxy domain and this helix is reputed to play a role in mediating interactions with other subunits. Crystal structures demonstrate that this terminal N helix buries its hydrophobic surface into an acceptor pocket of an adjacent Cre subunit[6].
The effect of the two domain structure is to form a C shaped clamp that grasps the DNA from opposite sides[3]
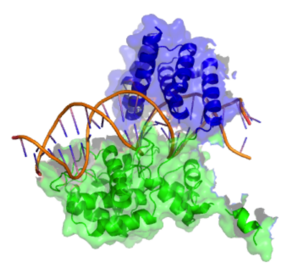

Active Site[edit]
The active site of the Cre enzyme consists of the conserved catalytic triad residues Arg 173, His 289, Arg 292 as well as the conserved nucleophilic residues Tyr 324 and Trp 315. Unlike some recombinase enzymes such as Flp Recombinase, Cre does not form a shared active site between separate subunits and all the reisdues that contribute to the active site are found on a single subunit. Consequently when two Cre molecules bind at a single loxP site two active sites are present. Cre mediated recombination requires the formation of a synapse in which two Cre-LoxP complexes associate to form what is known as the synapse tetramer in which 4 distinct active sites are present[6]. Tyr 324 acts as a nucleophile to form a covalent 3’-phosphotyrosine linkage to the DNA substrate. The scissile phosphate (phosphate targeted for nucleophilic attack at the cleavage site) is coordinated by the side chains of the 3 amino acid residues of the catalytic triad (Arg 173, His 289 & Trp 315). The indole nitrogen of tryptophan 315 also forms a hydrogen bond to this scissile phosphate. (n.b A Histidine occupies this site in other tyrosine recombinase family members and performs the same function). This reaction cleaves the DNA and frees a 5’ hydroxyl group. This process occurs in the active site of two out of the four recombinase subunits present at the synapse tetramer. If the 5’ hydroxyl groups attack the 3’-phosphotyrosine linkage one pair of the DNA strands will exchange to form a Holliday Junction intermediate[3].

Applications[edit]
Role in Bacteriophage P1[edit]
Cre Recombincase plays important roles in the life cycle of the P1 Bacteriophage. Upon infection of a cell the Cre-loxP system is used to cause circularization of the P1 DNA. In addition to this Cre is also used to resolve dimeric lysogenic P1 DNA that forms during the cell division of the phage[7].
Use in Research[edit]
The simplicity and robustness of the Cre-loxP systems has enabled scientists to exploit the Cre enzyme in order to manipulate DNA both in vivo and in vitro. See Cre-Lox Recombination for more details. The Cre enzyme can be expressed in many different organisms such as plants, bacteria, mammals, yeast. In 1992 Cre was expressed and found to be functional in a mouse host[8][9]. Promoter regions can be manipulated to allow precise temporal control of Cre enzyme expression (Eg. RU486-sensitive chimeric regulator GLVP[10]. As the enzyme has a specific 34bp DNA substrate the genome of the organism would have to be 1018 bp in lenght for there to be a likely occuranace of a loxP site. As mammalian genomes are on average in the region of 3x109 bp there is a very low chance of finding an endogenous loxP site[1]. For Cre to be fucntional in a foreign host, exogenous loxP sites must be engineereed. This allows precise control over the actvity of the Cre enzyme in test organisms.
References[edit]
- ^ a b Nagy A (2000). "Cre Recombinase:The Universal Reagent for Genome Tailoring". Genesis. 26: 99–109. PMID 10686599.
- ^ a b c d Abremski & Hoess (1984). "Bacteriophage P1 Site Specific Recombination. Purification and Properties of the Cre Recombinase Protein". Journal of Biological Chemistry. 259: 1509–1514. PMID 6319400.
- ^ a b c d Van Duyne G (2001). "A Strucutral View of Cre-loxP Site Specific Recombination". Annual Reviews Biophysics Biomolcular Strucutres. 30: 87–104. PMID 11340053.
- ^ >Ennifar E, Meyer J, Buchholz F, Stewart A & Suck D (2003). "Crystal strucutre of a wild type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation". Nucleic Acids Research. 31: 5449–5460. doi:10.1093/nar/gkg732.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sternberg & Hamilton (1981). "Bacteriophage P1 site-specific recombination:1.Recombination between loxP sites". Journal of Molecular Biology. 150: 467–486. doi:10.1016/0022-2836(81)90375-2.
- ^ a b Guo F, Gopaul D & Van Duyne G (1997). "Structure of Cre recombinase complexed with DNA in a site-specific recombiantion synapse". Nature. 389: 40–46. doi:10.1038/37925.
- ^ Shaikh & Sadowski (1997). "The Cre Recombinase Cleaves the Lox Site in Trans". [The Journal of Biological Chemistry]. 272: 5695–5702. doi:10.1074/jbc.272.9.5695.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Lakso M, Sauer B, Mosinger B, Lee E, Manning R, Yu S, Mulder K, Westphal H (1992). "Targeted oncogene activation by site specific recombination in transgenic mice". Proceedings of the National Academy of Sciences of the United States of America. 89: 6232–6236. PMID 1631115.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Orban p, Chui D, Marth JD (1992). "Tissue and site specific DNA recombination in transgenic mice". Proceedings of the National Academy of Sciences of the United States of America. 89: 6861–6865. PMID 1495975.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kyrkanides S, Miller J, Bowers W, Federoff H (2003). "Transcriptional and Posttranslational Regulation of Cre Recombinase by RU486 as the basis for an Enhanced Inducible Expression System". Nature Molecular Therapy. 8: 790–795. doi:10.1016/j.ymthe.2003.07.005.
{{cite journal}}: CS1 maint: multiple names: authors list (link)

